Introduction
do_x3dna has been developed for analysis of the DNA/RNA dynamics during the molecular dynamics simulations. It consists of three main components:
do_x3dna — To calculate structural descriptors of DNA/RNA from MD trajectory. Also available as a stand-alone and independent VMD plugin.
dnaMD — Command line tool to extract and analyze the data obtained from do_x3dna for users without programming experiences.
dnaMD Python module — To extract and analyze the data obtained from do_x3dna for users with programming experiences.
Warning
DSSR-X3DNA is incompatible with do_x3dna. Please use original 3DNA packacge. This could be dowloaded from by 3DNA forum.
Last Update: Nov. 2023
For Questions and Discussions, please visit: do_x3dna forum.
Release Note 2023
do_x3dna can be compiled and used with GROMACS-2023.x versions.
Support for older GROMACS (4.5.x, 4.6.x, 5.0.x, 5.1.x, 2016.x and 2018.x) is removed.
For above listed GROMACS version, the code is available in 2018 git branch. Please use
git checkout 2018to use this version.
Release Note 2018
Added support to calculate both global and local Elastic properties
Added tutorials to calculate elastic properties and deformation energy using dnaMD tool and dnaMD Python module.
Release Note 2017
do_x3dna can be compiled and used with GROMACS 4.5.x, 4.6.x, 5.0.x, 5.1.x, and 2016.x versions.
More user friendly — dnaMD tools to analyze do_x3dna data — No programming experiences needed now to analyze do_x3dna data.
Features
Base-pair parameters
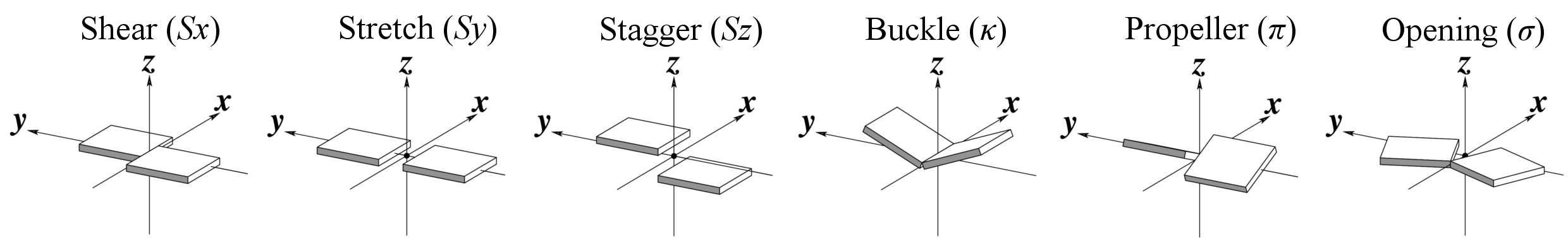
Base-step parameters
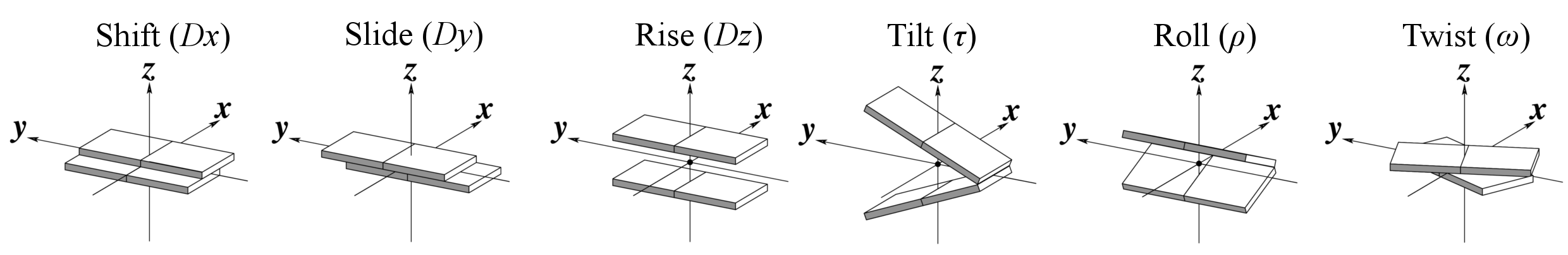
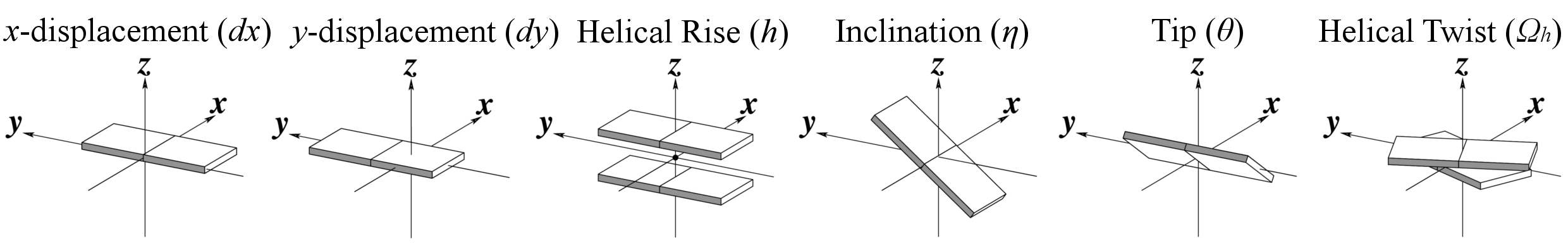
Helical Base-step parameters
Major and minor grooves
Radius of DNA Helix
Backbone torsional angles
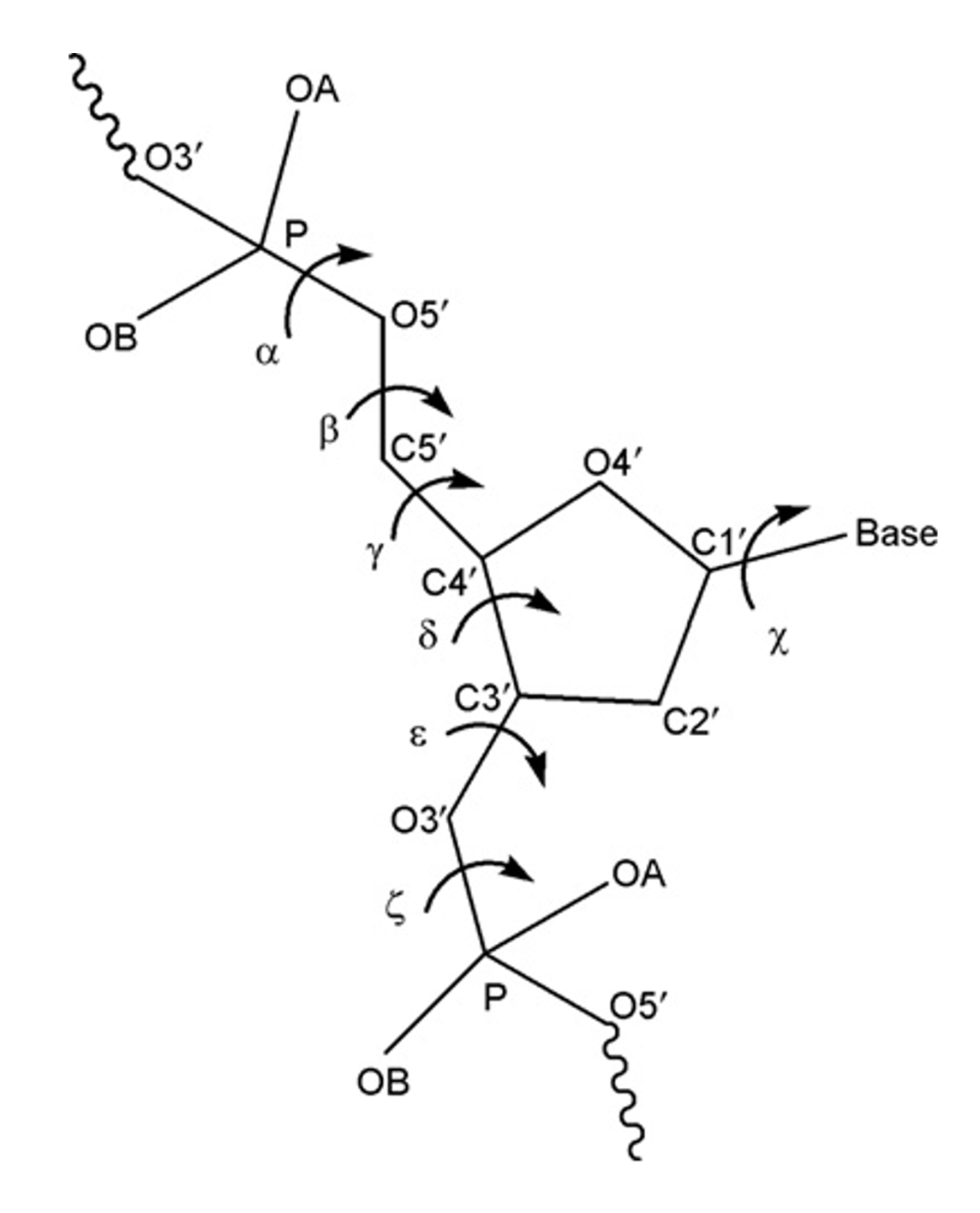
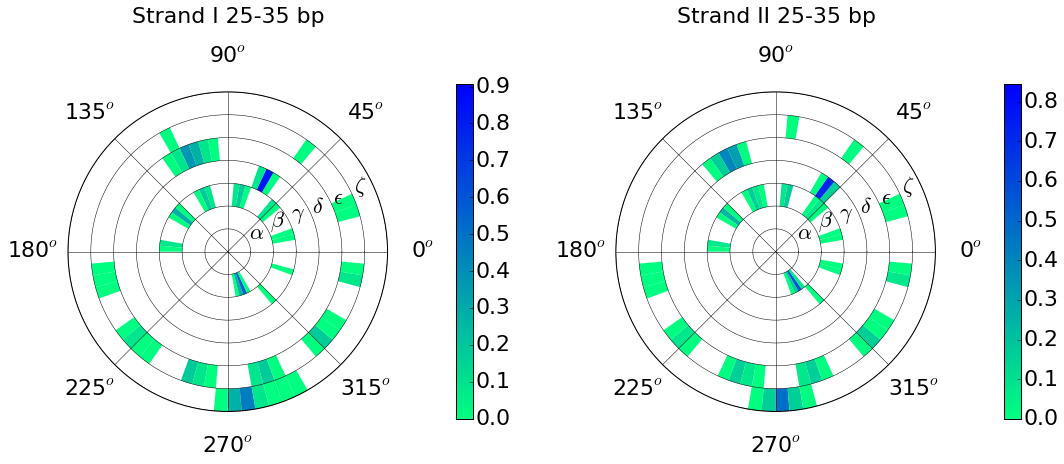
Example: Conformational wheel using backbone torsion angles:
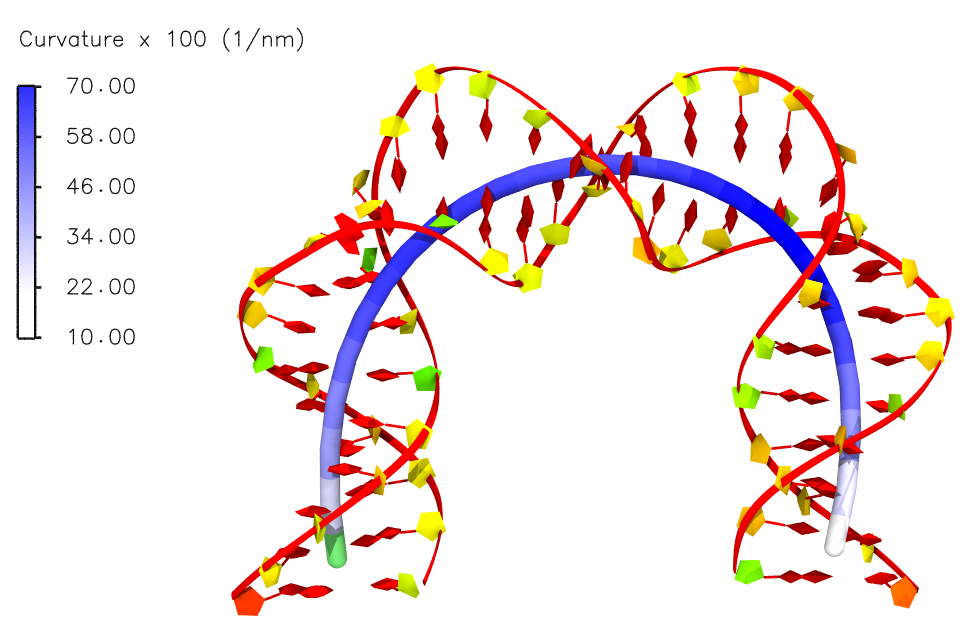
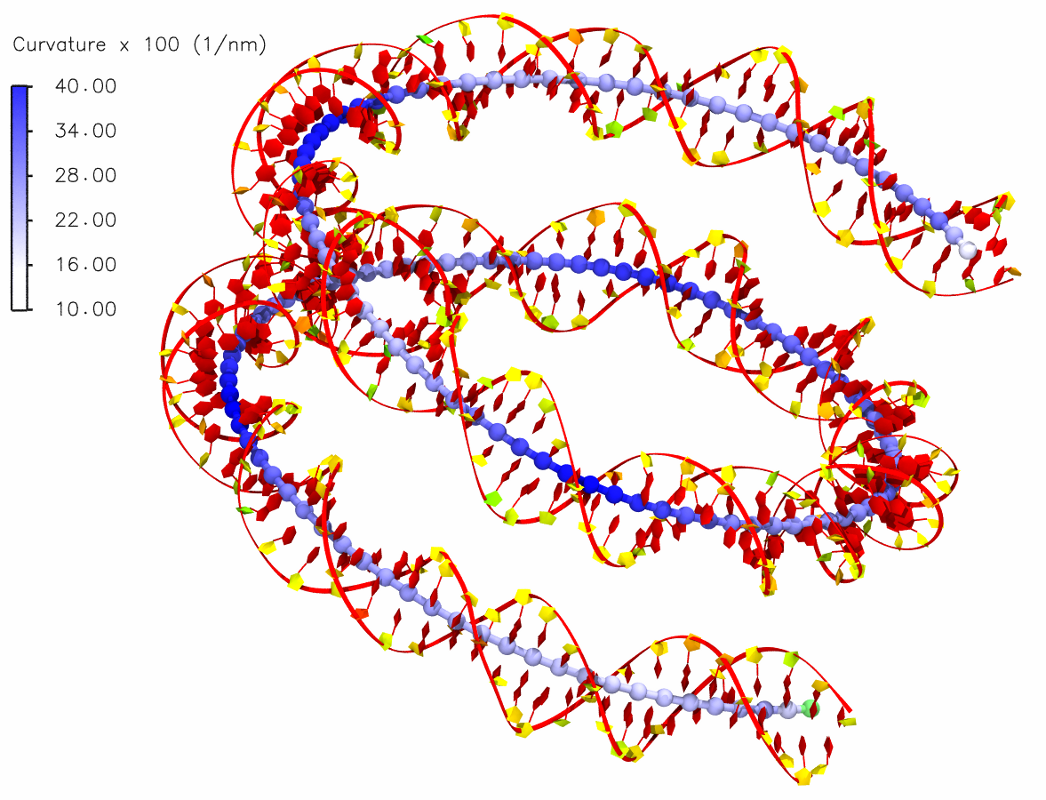
Helical axis and its curvature
Examples:
Bending in the helical axis of a mitochondrial DNA:
Bending in the helical axis of a DNA in the nucleosome:
Citations
Please cite the following publications:
- Xiang-Jun Lu & Wilma K. Olson (2003)3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures.Nucleic Acids Res. 31(17), 5108-21.
- Rajendra Kumar and Helmut Grubmüller (2015)Bioinformatics (2015) 31 (15): 2583-2585.